D Other datasets
D.1 BARCELONA dataset
All patients with an established diagnosis of IBD, including Crohn’s disease, ulcerative colitis, unclassified IBD, indeterminate colitis, or pouchitis, who were starting anti-TNF therapy with a biologic agent were monitored, in accordance with scheduled clinical visits, laboratory tests, imaging procedures and biologic sampling. This continued at 14 weeks and 46 weeks when a biopsy was taken during an ileocolonoscopy. This protocol was approved by the Institutional Ethics Committee of the Hospital Clinic de Barcelona (Study Number HCB/2012/7845 and HCB/2012/7956).
Those patients who had already started treatment with a biological agent at another center who were referred to Hospital Clínic de Barcelona’s IBD unit, were also included on the study, adapting to the corresponding time-schedule of their treatments. For all patients, starting anti-tumor necrosis factor (TNF) treatment was decided prior the protocol entry decision, according to best clinical practice.
Anonymized identification of the patients, disease, sex age at diagnosis, age at the time of the sample taking, time since the start of the treatment and sample segment was collected.
| Characteristic | BARCELONA |
|---|---|
| Individuals | 62 |
| Status (non-IBD/CD/UC) | 8/33/21 |
| Sex (female/male) | 29/33 |
| Age at diagnosis (<17/<40/>40 years) | 2/44/8 |
| Years of disease: mean (min-max) | 7.6 (0-32) |
| Age: mean (min-max) | 41 (18-68) |
| Time (0/14/46 weeks) | 41/40/32 |
| Sample segment (ileum/colon) | 39/87 |
The process of DNA extraction and sequencing was different for this dataset. We used different 16S-V3V4 primers (pair 341f/806r) on a MiSeq Nano sequencing as provided by the RTSF Genomics Core at Michigan State University, United State of America. The sequence of the primers used was as follows:
341f: 5’-CCTACGGGAGGCAGCAG-3’
806r: 5’-GGACTACHVHHHTWTCTAAT-3’
The results of the MiSeq Nano were processed with bcl2fastq (v1.8.4).
This dataset was processed as is usually done, although as part of the quality controls for the dataset the diversity measures of the samples was analyzed and displayed in figure D.1:
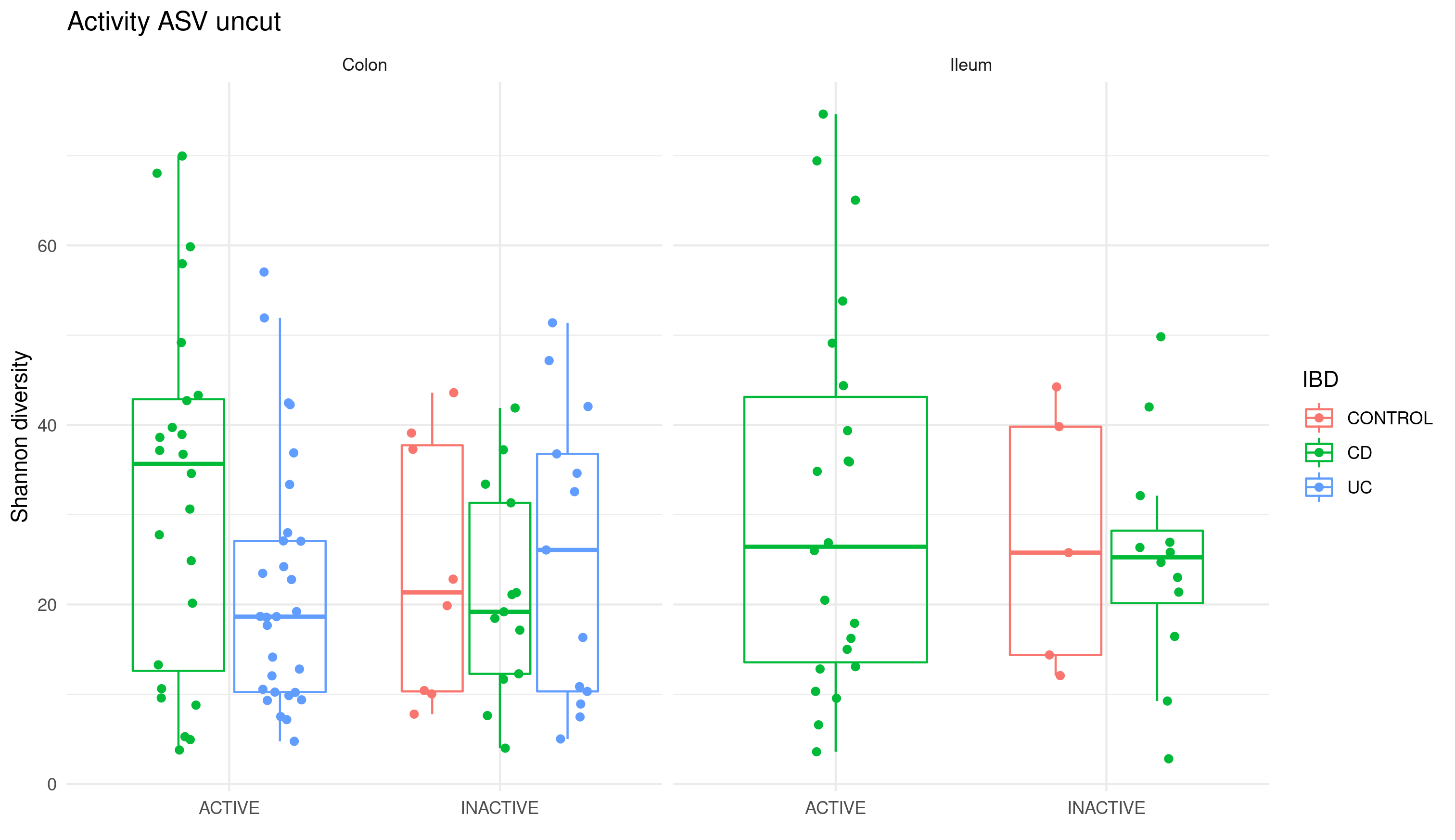
Figure D.1: Diversity indices of the BARCELONA cohort based on the location and disease status. There was considerable diversity among the different groups; however, importantly, the control samples overlapped with those patients presenting inflammatory bowel disease.
Control sample diversity should have been lower and not within the same range as those samples from patients with IBD. The dataset’s 16S was sequenced several times via different platforms. Despite the pilots and the negative controls used during the sequencing process, various problems appear each time: contamination, low quality and then this suspicious diversity issue. As it does not appear to be a problem of the sequencing facility itself, this data was abandoned as unreliable.
D.2 Hernández’s dataset
This dataset was obtained from collaborators at Mount Sinai, Toronto, Canada [82].
Patients with UC or CD were recruited during regularly scheduled visits or via normal hospital surveillance protocols. In addition, asymptomatic healthy controls were recruited during routine, age-related colorectal cancer screening by colonoscopy. A total of 290 samples were collected together with information about the disease, age at diagnosis, age at the time of the sampling, sex, sample location and smoking status.
| Characteristics | Hernández |
|---|---|
| Disease (non-IBD/CD/UC) | 46/54/66 |
| Age at diagnosis (<17/<40/>40 years) | 29/73/18 |
| Age: mean (min-max) | 40 (17-71) |
| Sex (female/male) | 81/85 |
| Smoking (never/ex/current) | 115/34/16 |
| Sample Location (ileum/colon) | 97/193 |
D.2.1 Results
We substitutes the BARCELONA dataset (See section D.1) in order to confirm the results of the previous datasets. However, at the time of writing this the process remained incomplete.
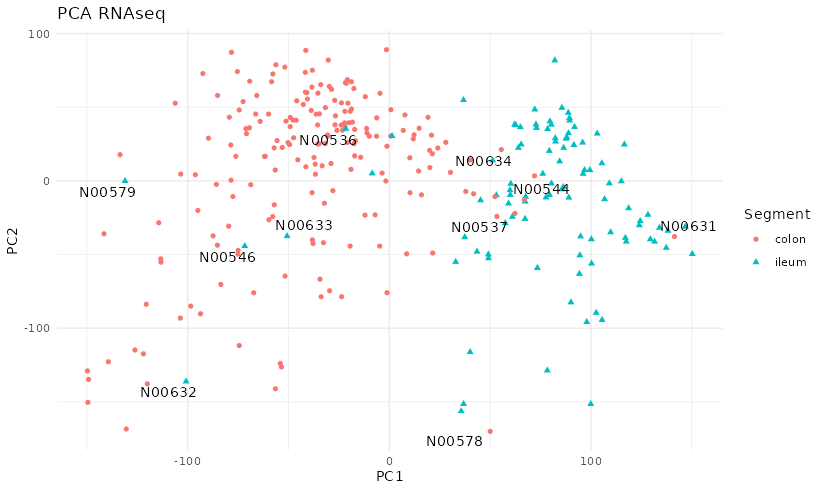
Figure D.2: PCA of RNAseq of the Hernandez’s dataset. The plot shows a clear separation between colon and ileum for most samples except for some that seem mislabeled.
There seems to be some samples that might be mislabeled as their corresponding RNA profile does not match those samples of the similar location. This could be from taking the sample from a region near a surgery, for which the expression profile might not match the usual patterns.